Polymerase Chain Reaction: Principles and Uses of PCR
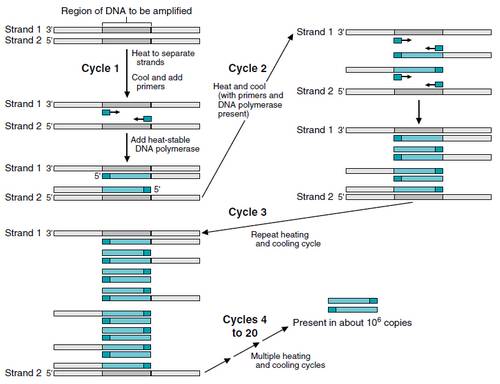
The Polymerase Chain Reaction (PCR) is an invitro method of DNA amplification that can rapidly clone (amplify) DNA samples as small as a single molecule. If a length of DNA is mixed with the 4 nucleotides (A, T, C and G), and the enzyme DNA polymerase, then the DNA will be replicated many times. Developed in 1983 by Kary Mullis, PCR is now a common technique used in medical and biological research labs for a variety of applications.
Components of a PCR reaction:
- A DNA template
- Heat stable Taq DNA polymerase (derived from the thermophilic bacterium Thermus aquaticus, which grows naturally in hot springs at a temperature of 90c, so is not denatured by the high temperatures)
- Nucleotides
- A pair of DNA primers
- Buffers containing magnesium ions
Comparison of PCR components with photocopier items
| Photocopier items | PCR components |
|---|---|
| 1. The book | DNA template |
| 2. The page | A portion of the genome (fragment) we are interested in |
| 3. A bookmark | Primers that “mark” the specific fragment |
| 4. Photocopier | DNA polymerase |
| 5. Paper and toner | The 4 nucleotides |
The principle of PCR is as follows:
1. Starting with a sample of the DNA (template) to be amplified, add the 4 nucleotides (deoxyribonucleoside triphosphates) and the enzyme heat stable DNA polymerase to the solution.
2. Normally (in vivo) the DNA double helix would be separated by the enzyme helicase, but in PCR (in vitro) the strands are separated by heating to 95 c for 2 minutes. This breaks the hydrogen bonds between the 2 DNA strands.
3. Initiation of DNA polymerisation always requires short lengths of DNA (about 20 bp long) called primers. The primers are 2 synthetic oligonucleiotides: one is complementary to a short sequence in one strand of the DNA to be amplified, and the other is complementary to a sequence in the other DNA strand. In vivo the primers are made during replication by DNA polymerase, but in vitro they must be synthesized separately and added at this stage. This means that a short length of the sequence of the DNA must already be known.
4. The DNA must be cooled to 40 c to allow the primers to anneal to their complementary sequences on the separated DNA strands.
5. The DNA polymerase enzyme can now extend the primers and complete the replication of the rest of the DNA. Its optimum temperature is about 72 c, so the mixture is heated to this temperature for a few minutes to allow replication to take place as quickly as possible.
5. Each original DNA molecule has now been replicated to form 2 molecules. The cycle is repeated from step 2, each time doubling the number of DNA molecules. This is why it is called a chain reaction, since the number of molecules increases exponentially. Typically PCR is run for 20–30 cycles.
PCR can be completely automated; a minute sample of DNA can be amplified millions of times with little effort.
 Description of the schematic diagram for PCR technique: Strand 1 and Strand 2 are original DNA strands. The short dark blue fragments are the primers. After multiple heating and cooling cycles, the original strands remain, but most of the DNA consists of amplified copies of the segment (shown in lighter blue) synthesized by the heat stable DNA polymerase.
Description of the schematic diagram for PCR technique: Strand 1 and Strand 2 are original DNA strands. The short dark blue fragments are the primers. After multiple heating and cooling cycles, the original strands remain, but most of the DNA consists of amplified copies of the segment (shown in lighter blue) synthesized by the heat stable DNA polymerase.
Uses of PCR:
1. It is appropriate for forensic testing procedures because only a very small sample of DNA is required as the starting material which can be obtained from a single strand of hair or a single drop of blood or semen.
2. Tissue typing for organ transplantation
3. DNA based phylogeny or functional analysis of genes useful for research
4. Detection of genetic diseases: Prenatal genetic diagnosis, Hereditary diseases
5. Detection of infectious diseases: HIV, TB, etc.
6. Parental testing, where an individual is matched with their close relatives
Disadvantage of PCR:
A potential problem for PCR is obtaining a pure sample of DNA to start with; any contaminant DNA will also be amplified.
Sources:
- Essential Biochemistry for Medicine
- Mark’s Medical Biochemistry

5 Comments
Can I get the exact syllabus for Basic science (MBBS Ist year) as per new curricula of Kathmandu University?
Siddhartha,
It is not possible to include entire KU MBBS syllabus here.
It is available in form of Book in the market. If we get a soft copy of it we will try to share it.
ok thanx.
This is a nice article. I would also recommend reading the article by biswajeet on Polymerase Chain Reaction.. He has covered the topic in more detail but for people who do not need the detail this article works well.
Thank you Akul.
Looks like a pretty well researched article.
Comments are closed.